The insect brain is built by a conserved set of modules, the neuropils. However, size, shape and development of these neuropils differ dramatically between species reflecting evolutionary adaptations. The brain forms from neural stem cells, the neuroblasts. However, it remains unknown what makes embryonic brain neuroblasts different from each other and what cellular and genetic mechanisms regulate evolutionary adaptations.
The evolution of the central complex
The central complex develops during postembryonic stages in Drosophila but during embryogenesis in hemimetabolous insects. Tribolium takes an intermediate position in that it develops the upper unit during embryogenesis and the lower unit postembryonically. We want to find out, what the cellular and genetic mechanisms are that govern this heterochronic shift of brain development.
To that end, we mark homologous neural lineages in Tribolium and Drosophila by genome editing and transgenesis and compare their development.
 |
|
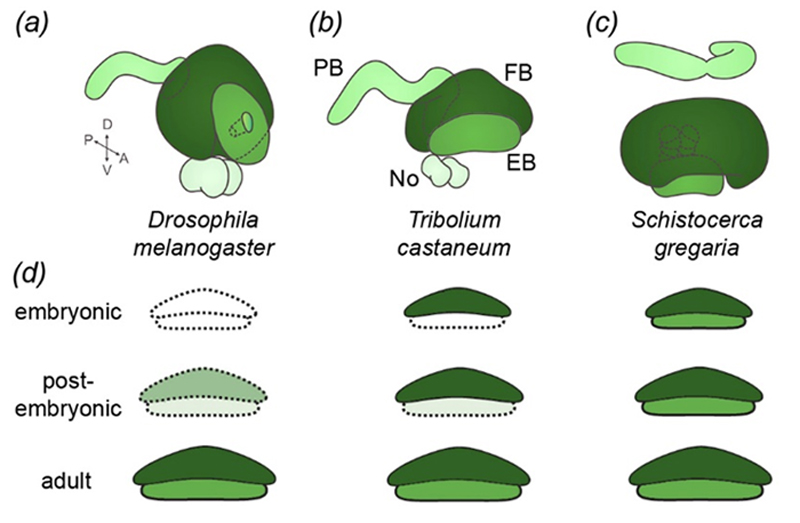
| (a-c) The central complex consists of the same modules in different species. (d) It develops during embryogenesis in hemimetabolous insects (Schistocerca) and postembryonically in Drosophila. Tribolium takes an intermediate position where the upper unit is formed during embryogenesis and the lower unit postembryonically. | |
The genetic control of embryonic central complex development
In Drosophila, the brain is built by about 200.000 neurons that arise from about 100 neural stem cells (neuroblasts, NBs) in each hemisphere. Before and/or shortly after delamination each NB can be identified by a unique code of expressed transcription factors and this code is likely to direct their developmental fate. The offspring of a given NB follow an invariant developmental path that is different for each NB. Therefore, the early determinants and signals that provide NBs with distinct identities are essential and instructive for the formation of a functional brain. These determinants and signals are well studied for the trunk but largely unknown for the brain neuroblasts.
To adress this question, we are developing imaging systems for marking neuroblast lineages, describe the brain structures and identify regions missing in RNAi knock-downs. Genes expressed in the neuroectoderm potentially constitute the signals for NB identity specification. By knocking down these genes by RNAi and by misexpression studies we try to understand the signals that make one NB different from the other and to understand the consequences on brain architecture.

